HuSSPred: Human Skin Sensitization Predictions
The application is a online web portal to predict skin sensitization potential, described in "Using ML for Human Skin Sensitization Prediction – a novel in silico Tool using HPPT data". To use, enter SMILES in the box below, or draw a compound and hit load SMILES, then click "Get Properties". Results will appear below. By default all models for all endpoints will be run. You can choose to turn off certain endpoints in the options sidebar. Fragment contribution maps are generated with RDKit. To turn on the maps, check the "Display contribution maps" in the options sidebar. It defaults to off because the maps will increase the runtime significantly, so if using please be patient. More information about these maps can be found here.
For the applicability domain calculation (AD), an ensemble confidence approach is used such that if the average prediction confidence of the ensemble of models is above 0.6, the prediction is considered "inside" the AD
Please cite "Using ML for Human Skin Sensitization Prediction – a novel in silico Tool using HPPT data". Models and code for this webserver can be found here.
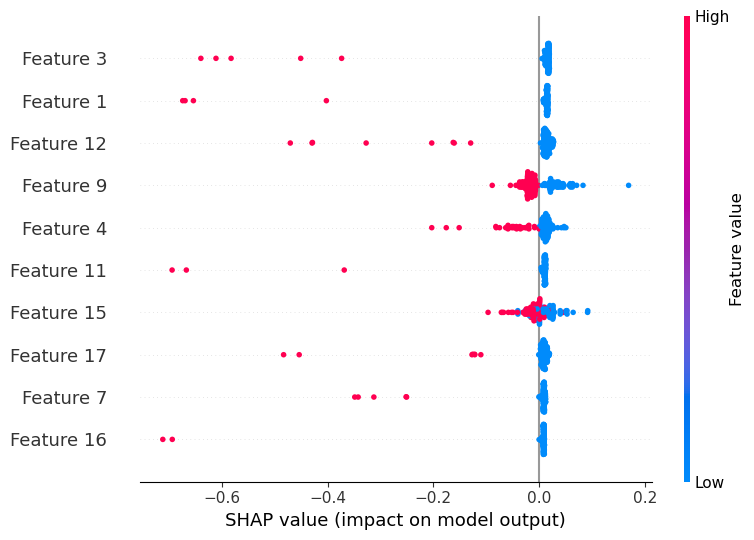
SHAP feature importance for predictive models
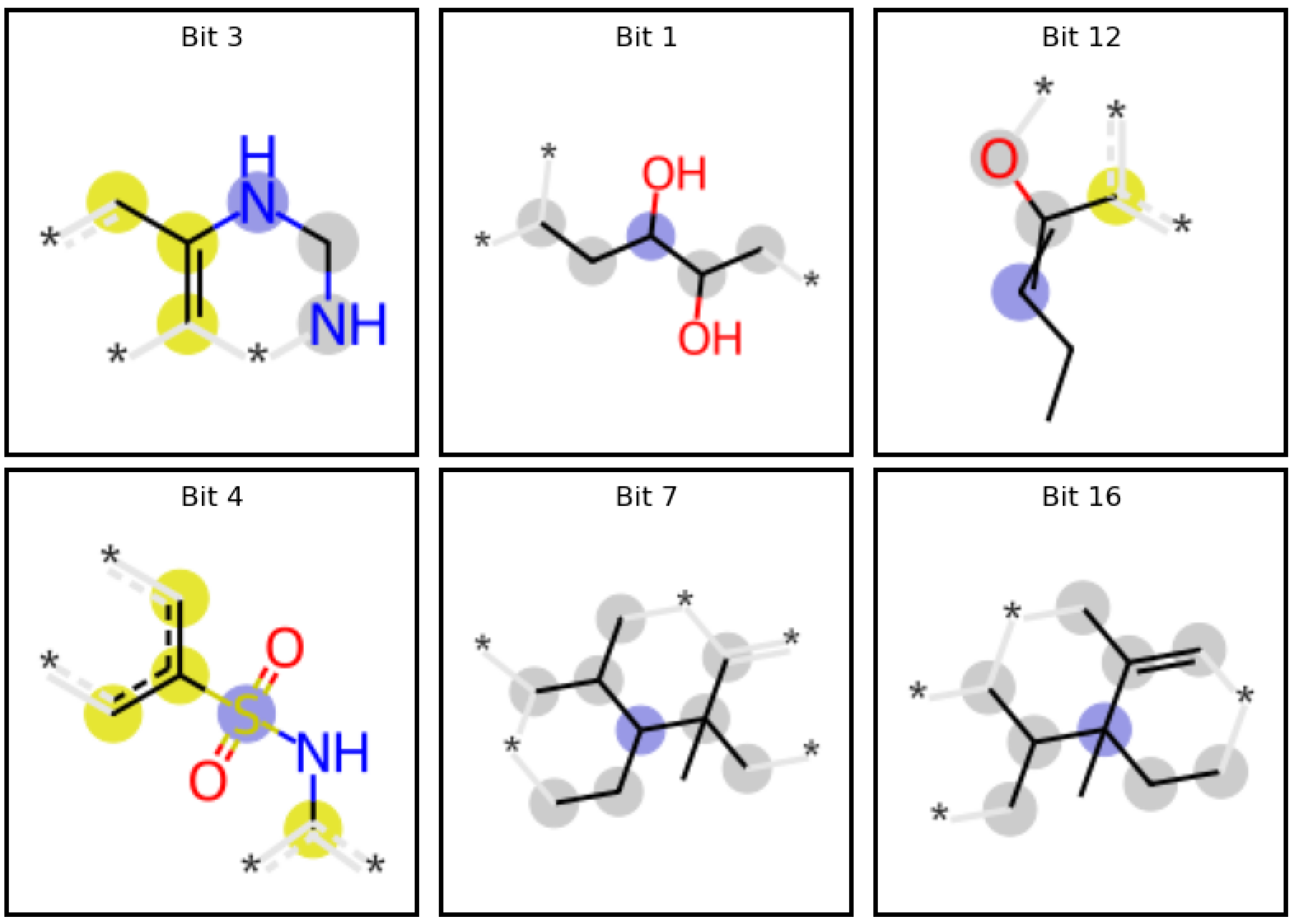

Results
Developed by James Wellnitz and Ricardo Tieghi of the MML @ UNC
Thank you to the NIEHS/NICEATM and the NSCP Program for the support. This work was supported in part by the NIH Intramural Research Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of National Institutes of Health
© 2025, all rights reserved